Understanding the Language of Life
Yanni SunDept. of Electrical Engineering City University of Hong Kong yannisun AT cityu DOT edu DOT hk [Publication] [Tools] [Opening] [Research Area][News][Students] |

|
Yanni Sun is a Professor in the Department of Electrical Engineering at City University of Hong Kong. Before she moved to Hong Kong, she was an Associate Professor in Computer Science and Engineering Department at Michigan State University, USA. She received the B.S. and M.S. degrees from Xi’an JiaoTong University (China), both in Computer Science. She received the Ph.D. degree in Computer Science from Washington University in Saint Louis, USA. Her research field is bioinformatics and computational biology. In particular, her recent research projects include applying deep learning models in biological sequence analysis, next-generation sequencing data analysis, metagenomics, protein domain annotation, plant genomics, and noncoding RNA annotation. She was a recipient of NSF CAREER Award in 2010.
Education
-
Ph.D. in Computer Science, Washington University in St. Louis, MO, USA
-
Thesis: Designing Filtration Strategies for Fast Sequence Annotation
-
Advisor: Dr. Jeremy Buhler (Advisory committee: Dr. Jeremy Buhler, Dr. Gary Stormo, Dr. Michael Brent, Dr. Sally Goldman, Dr. Tao Ju, and Dr. Nan Lin).
-
M.S. and B.S. in Computer Science, Xi’an JiaoTong University, Xi’an, China
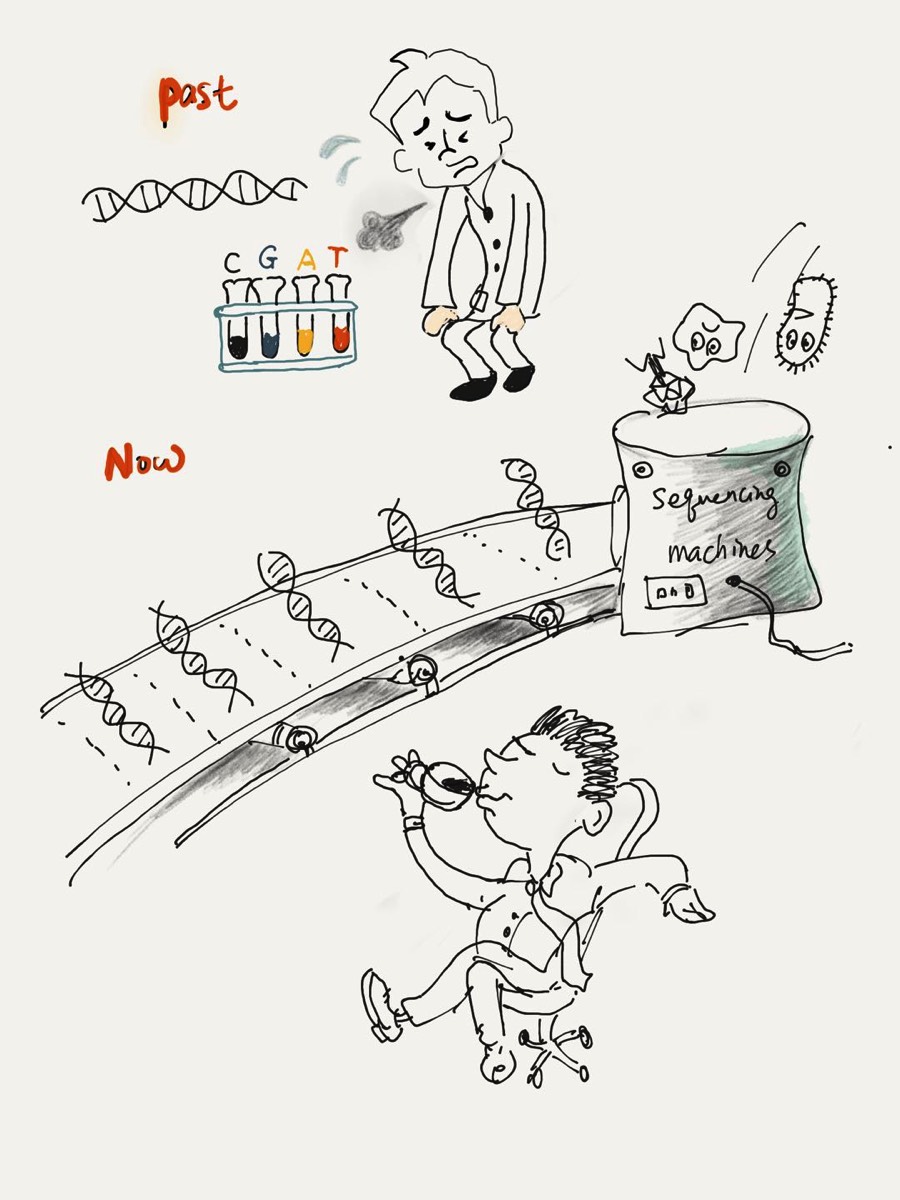
Research Interests
|

|
- Computational biology and Bioinformatics
- deep learning-based sequence analysis
- BIG genomic data mining
- algorithm design and tool development
- Microbial community analysis using high-throughput sequencing data
- Composition analysis of metagenomics data sequenced from host-associated or environmental samples
- Gene-centric functional anlaysis of metagenomic data
- Virus diversity analysis using deep sequencing data
- Viral population characterization using shot-gun deep sequencing data
- Clinically important virus (such as HIV, HCV, SARS-CoV) identification
- Virus identification, classification, and host prediction using deep learning models
- Plant genome annotation
- Gene prediction in crops such as rice and maize
- Small nocoding RNA search in plant genomes
- Algorithm design for mining third-generation sequencing data
- Error correction of long reads
- De novo assembly of virus genomes using long reads
- Noncoding RNA (small RNA, lncRNA etc.) secondary structure identification and search in large-scale genomic databases
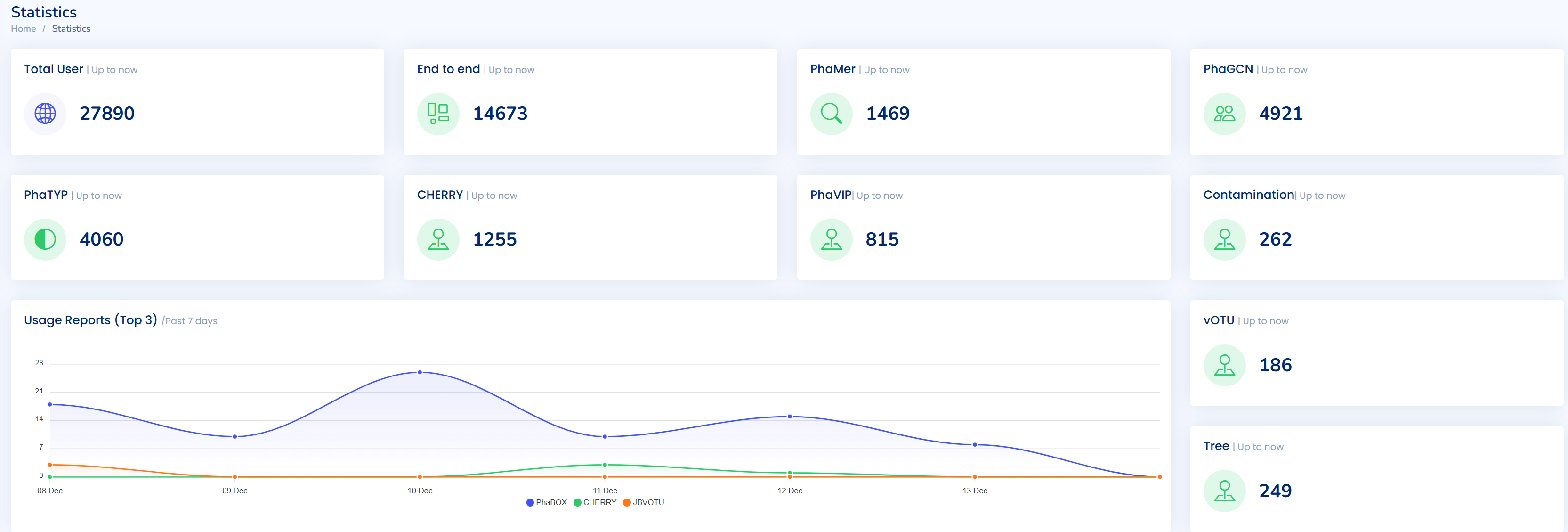
WebServer for Virus Analysis: Phabox2

|
|